Part III of The New Genetics
If we are to understand the role of genes within a living organism, the first thing we must understand is that genes, by themselves, are inert. Put a gene in a petri dish with all the nutrients needed to make a protein and it will do nothing. Nada. Genes do not express themselves. Cells express genes.
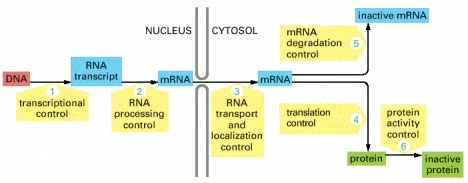
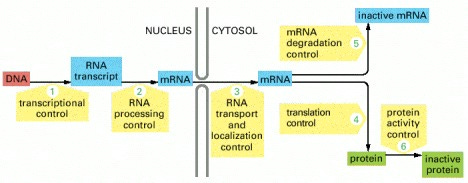
A eukaryotic cell — a cell from an organism of higher complexity than bacteria — must use hundreds of proteins to make a single functional protein from a gene. Here is a schematic showing the different steps a cell takes to make a functional protein:
Image hosted at pubmed's searchable but not browsable version of Molecular Biology of the Cell.
The cell can regulate each step in response to its own needs and the needs of the organism of which it is part, constantly adjusting the expression of its genes to fit its environment.
What follows is a brief summary of chapter six of Molecular Biology of the Cell, the definitive guide to mainstream molecular biology.
The first thing the cell must do is make an RNA transcript called messenger RNA or mRNA that will carry the information from the DNA within the nucleus to the cellular fluid that lies outside, called the cytosol. This process is called transcription because the code is staying within the “language” of nucleic acid, but changing form somewhat, like converting a handwritten message into a typed message.
Although RNA polymerase is the enzyme that assembles the mRNA, this process generally requires well over a hundred proteins to get going.
Once the polymerase begins transcribing the gene, it sheds this first assembly of proteins, called transcription factors, and begins associating with a number of proteins called elongation factors required for it to keep moving along.
Before the nucleus exports the mRNA, the mRNA must be edited. The splicing machinery required for this process involves five RNA-based enzymes and as many as 200 additional proteins.
The mRNA must also have a cap made from a modified form of guanine, one of the nucleotides that makes up the basic structure of DNA. It takes three enzymes to make the cap. Several other sets of proteins are required to give the other end of the mRNA transcript a poly-A tail made of a string of adenine nucleotides. Adenine is another basic building block of DNA like guanine. The cap and tail of the mRNA help identify it as a transcript destined for protein synthesis and provide the cell with several additional mechanisms to control the amount of protein it makes from the transcript, as discussed in more detail in the next post in this series.
The RNA processing enzymes are generally seen to be stockpiled in granules poised nearby transcription sites, ready for use. But when transcription begins, they become tethered to a long tail protruding from RNA polymerase, so Molecular Biology of the Cell calls the polymerase an “RNA factory.”
Much of the spliced material is discarded, and the nucleus uses specific receptors and nuclear pore complexes to transport the usable mRNAs into the cytosol. In order to make proteins out of it, however, the cell must have protein-making machines made primarily of RNA called ribosomes. A growing mammalian cell contains ten million ribosomes.
The ribosome is a ribozyme, a word used to describe enzymes that are made from RNA instead of protein. While four different RNA molecules make up two thirds of the ribosome, each ribosome also has fifty different proteins that lock into the holes and crevices in its surface, where they appear to stabilize its shape and facilitate its movement.
In order to make enough RNA to manufacture millions of ribosomes, multiple copies of the four unique RNA-encoding genes are required. Humans therefore have 200 genes located on five chromosomes that code for ribosomal RNA.
In order to actually assemble each ribosome, 150 different “guide RNAs” are required to make approximately 200 specific chemical modifications to the ribosome. This occurs in a structure within the nucleus called the nucleolus, which Molecular Biology of the Cell calls a “ribosome factory.”
While the cell's ten million ribosomes are primarily involved in using mRNA transcripts to make proteins, the first role of the ribosome is to enforce quality control (QC) on the mRNA transcripts as they exit the nucleus. During the mRNA splicing we discussed above, each splice site is tagged with special proteins. As the mRNA exits the nucleus, a special ribosome examines it to make sure the “stop” message occurs after the last of these labels. If not, the QC managing ribosome will identify it as an mRNA that was either incorrectly spliced, or produced from a defective gene, and will target the mRNA for destruction. If so, the QC manager removes the labels and allows the transcript into the cytosol where it can be used to make proteins.
The process of making a protein from the mRNA transcript is called translation, because it is translating the message from the language of nucleic acid to the language of protein.
Several sets of proteins are involved in carrying the amino acids to the ribosome in a way that ensures accurate reading of the genetic code. There are 48 unique transfer RNAs (tRNAs) encoded in multiple copies by 500 genes. These tRNAs are shaped like a cross or a three-leaf clover and attach an amino acid on one end, and read the genetic code of the mRNA transcript on the other end.
There are 20 aminoacyl-tRNA synthetases, which are enzymes that attach the amino acids to the proper tRNA molecules. These enzymes also contain an “editing” capacity that fixes mistakes where the wrong amino acid gets joined to the tRNA molecule.
Finally, several elongation factors are also required to increase the speed and accuracy of protein production at the site of the ribosome.
The protein exits the ribosome through a tunnel in a mostly unfolded, linear state. Its final functional properties, however, depend on its three-dimensional structure after it has folded. The information for the three-dimensional structure is partly contained in the linear sequence of amino acids, but not always entirely.
As the protein exits through the ribosomal tunnel, chaperone proteins often help it fold properly. Once it has exited, other chaperone proteins sometimes form a barrel-shaped “isolation chamber” around the protein to help it finish folding, or to help misfolded proteins refold. There are several major families of chaperone proteins, and they are often specific to certain proteins and even to certain organelles within the cell.
Alternative ways of folding a protein are most commonly known in disease states, such as in the prion theory of mad cow disease. However, alternative protein folding states have been shown to contribute to the formation of different cell types in certain fungi, and it is possible that protein folding could represent a form of non-genetic inheritance, as discussed in more general terms in the second post in this series.
Some researchers have made the controversial suggestion that alternative forms of protein folding could contribute to memory in complex organisms such as humans.
Once the final protein is folded, the cell still has many ways of modifying the protein to make it active, inactive, or to change how it functions or just how active it is.
This has been, of course, a simple summary of what is contained in Molecular Biology of the Cell. Despite all of the complexity that has been discovered, the authors still state that “an enormous amount of ignorance remains; many fascinating challenges therefore await the next generation of cell biologists.”
What we do know, however, should make it clear that genes do not express themselves. It should, by now, be abundantly clear that cells express genes.
In the next post, we will take a look at the fascinating way in which cells control the expression of their genes in response to their environment, in ways that fulfill their own needs and the needs of the organism of which they are part. Now that we've covered some of the basics, things will really get interesting. Stay tuned.